stanflow is currently under active development–breaking changes may be made without warning, until a proper release.
stanflow offers an integrated, mildly opinionated access to a Stan-based Bayesian Workflow (Gelman et al. 2020). Much like the famous tidyverse package, stanflow is a metapackage which installs and attaches relevant Stan R packages, serving as a one-stop-shop for Bayesian modelling. stanflow draws heavy inspiration from tidyverse, and reuses portions of its codebase–however, stanflow eschews tidyverse packages (i.e., purrr, dplyr, etc.) for base R whenever necessary.
Installation
You can install the development version of stanflow from GitHub with:
# install.packages("pak")
pak::pak("VisruthSK/stanflow")Usage
Here, we load stanflow and decide to use cmdstanr as the backend for brms.
library(stanflow)
#> ── Attaching Stan processing packages ─────────────────── stanflow 0.0.0.9000 ──
#> ✔ bayesplot 1.15.0.9000 ✔ projpred 2.10.0
#> ✔ loo 2.9.0 ✔ shinystan 2.7.0
#> ✔ posterior 1.6.1
#> ── Available Stan interfaces ────────────────────────────── setup_interface() ──
#> • brms 2.23.0 • rstan 2.32.7
#> • cmdstanr 0.9.0 • rstanarm 2.32.2
#> ── Conflicts ─────────────────────────────────────────── stanflow_conflicts() ──
#> ✖ posterior::rhat() masks bayesplot::rhat()
#> ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
setup_interface(
interface = "brms",
dev = FALSE,
brms_backend = "cmdstanr",
cores = 2,
quiet = FALSE,
force = FALSE,
check_updates = TRUE
)
#> ℹ Adding cmdstanr to setup because `brms_backend = 'cmdstanr'`
#> ℹ Attaching brms...
#> ℹ Configured brms: set `options(mc.cores = 2)` and `options(brms.backend = 'cmdstanr')`
#> ℹ Attaching cmdstanr...
#> The C++ toolchain required for CmdStan is setup properly!
#> ℹ Found CmdStan v2.37.0 at 'C:/Users/visru/.cmdstan/cmdstan-2.37.0'
#> ! Update available: v2.37.0 -> v2.38.0
#> ℹ Skipping update in non-interactive mode (set `force = TRUE` to upgrade).
#> ✔ Setup complete. brms, cmdstanr packages are attached; you do not need to run `library()`.
set.seed(0)
flow_check()
#> ── Attaching Stan processing packages ─────────────────── stanflow 0.0.0.9000 ──
#> ✔ bayesplot 1.15.0.9000 ✔ projpred 2.10.0
#> ✔ loo 2.9.0 ✔ shinystan 2.7.0
#> ✔ posterior 1.6.1
#> ── Available Stan interfaces ────────────────────────────── setup_interface() ──
#> ✔ brms 2.23.0 • rstan 2.32.7
#> ✔ cmdstanr 0.9.0 • rstanarm 2.32.2
#> ── Conflicts ─────────────────────────────────────────── stanflow_conflicts() ──
#> ✖ brms::ar() masks stats::ar()
#> ✖ brms::do_call() masks projpred::do_call()
#> ✖ brms::rhat() masks posterior::rhat(), bayesplot::rhat()
#> ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsWith the first two commands (note that the setup_interface() call is unnecessarily verbose), users have access to everything they need to fit and interrogate Bayesian models. Specifically, those two commands will load the core stanflow packages: bayesplot, loo, posterior, projpred and shinystan, as well as setup brms to use the cmdstanr backend with as many cores as are available on your machine. These package (inlcuding brms and cmdstanr) are all “library()d” as well, so we can proceed with our analysis immediately.
cmdstanr_example()
#> variable mean median sd mad q5 q95 rhat ess_bulk ess_tail
#> lp__ -65.94 -65.62 1.39 1.19 -68.67 -64.29 1.00 1996 2876
#> alpha 0.38 0.37 0.21 0.22 0.03 0.74 1.00 4087 3110
#> beta[1] -0.67 -0.67 0.25 0.25 -1.09 -0.27 1.00 4516 2758
#> beta[2] -0.27 -0.26 0.22 0.22 -0.64 0.09 1.00 3520 3012
#> beta[3] 0.67 0.66 0.27 0.27 0.24 1.12 1.00 4444 3098
#> log_lik[1] -0.51 -0.51 0.10 0.10 -0.68 -0.37 1.00 3705 3254
#> log_lik[2] -0.41 -0.39 0.15 0.14 -0.67 -0.20 1.00 4327 3192
#> log_lik[3] -0.50 -0.46 0.22 0.20 -0.91 -0.20 1.00 3433 2935
#> log_lik[4] -0.45 -0.43 0.15 0.15 -0.73 -0.24 1.00 3605 2931
#> log_lik[5] -1.18 -1.16 0.28 0.28 -1.67 -0.75 1.00 4895 3413
#>
#> # showing 10 of 105 rows (change via 'max_rows' argument or 'cmdstanr_max_rows' option)
fit1 <- brm(
count ~ zAge + zBase * Trt + (1 | patient),
data = epilepsy,
family = poisson(),
silent = TRUE,
refresh = 0
)
#> Start sampling
#> Running MCMC with 4 chains, at most 2 in parallel...
#>
#> Chain 2 finished in 7.0 seconds.
#> Chain 1 finished in 7.0 seconds.
#> Chain 3 finished in 7.0 seconds.
#> Chain 4 finished in 7.4 seconds.
#>
#> All 4 chains finished successfully.
#> Mean chain execution time: 7.1 seconds.
#> Total execution time: 14.7 seconds.
summary(fit1)
#> Family: poisson
#> Links: mu = log
#> Formula: count ~ zAge + zBase * Trt + (1 | patient)
#> Data: epilepsy (Number of observations: 236)
#> Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
#> total post-warmup draws = 4000
#>
#> Multilevel Hyperparameters:
#> ~patient (Number of levels: 59)
#> Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
#> sd(Intercept) 0.58 0.07 0.46 0.74 1.01 751 1459
#>
#> Regression Coefficients:
#> Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
#> Intercept 1.76 0.12 1.53 1.99 1.01 728 1469
#> zAge 0.09 0.08 -0.07 0.26 1.00 848 1597
#> zBase 0.70 0.12 0.46 0.94 1.00 816 1347
#> Trt1 -0.26 0.17 -0.57 0.07 1.00 714 1196
#> zBase:Trt1 0.05 0.17 -0.28 0.38 1.00 904 1528
#>
#> Draws were sampled using sample(hmc). For each parameter, Bulk_ESS
#> and Tail_ESS are effective sample size measures, and Rhat is the potential
#> scale reduction factor on split chains (at convergence, Rhat = 1).
plot(fit1, variable = c("b_Trt1", "b_zBase"))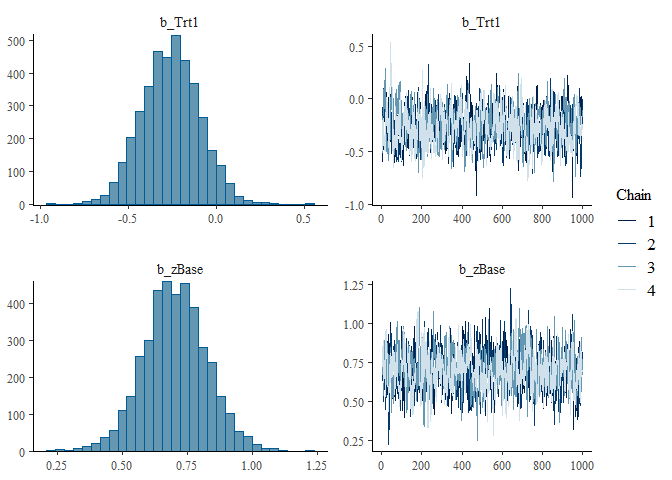
fit2 <- brm(
count ~ zAge + zBase * Trt + (1 | patient) + (1 | obs),
data = epilepsy,
family = poisson(),
silent = TRUE,
refresh = 0
)
#> Start sampling
#> Running MCMC with 4 chains, at most 2 in parallel...
#>
#> Chain 1 finished in 11.3 seconds.
#> Chain 2 finished in 11.5 seconds.
#> Chain 4 finished in 13.6 seconds.
#> Chain 3 finished in 14.8 seconds.
#>
#> All 4 chains finished successfully.
#> Mean chain execution time: 12.8 seconds.
#> Total execution time: 26.3 seconds.
summary(fit2)
#> Family: poisson
#> Links: mu = log
#> Formula: count ~ zAge + zBase * Trt + (1 | patient) + (1 | obs)
#> Data: epilepsy (Number of observations: 236)
#> Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
#> total post-warmup draws = 4000
#>
#> Multilevel Hyperparameters:
#> ~obs (Number of levels: 236)
#> Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
#> sd(Intercept) 0.37 0.04 0.29 0.46 1.00 1349 1967
#>
#> ~patient (Number of levels: 59)
#> Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
#> sd(Intercept) 0.54 0.07 0.41 0.70 1.00 1125 1797
#>
#> Regression Coefficients:
#> Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
#> Intercept 1.72 0.12 1.49 1.95 1.00 1088 2092
#> zAge 0.09 0.09 -0.09 0.26 1.00 1055 1569
#> zBase 0.70 0.12 0.47 0.94 1.00 971 1557
#> Trt1 -0.26 0.16 -0.59 0.06 1.00 922 1751
#> zBase:Trt1 0.06 0.16 -0.26 0.37 1.00 1045 1803
#>
#> Draws were sampled using sample(hmc). For each parameter, Bulk_ESS
#> and Tail_ESS are effective sample size measures, and Rhat is the potential
#> scale reduction factor on split chains (at convergence, Rhat = 1).
loo(fit1, fit2)
#> Warning: Found 6 observations with a pareto_k > 0.7 in model 'fit1'. We
#> recommend to set 'moment_match = TRUE' in order to perform moment matching for
#> problematic observations.
#> Warning: Found 58 observations with a pareto_k > 0.7 in model 'fit2'. We
#> recommend to set 'moment_match = TRUE' in order to perform moment matching for
#> problematic observations.
#> Output of model 'fit1':
#>
#> Computed from 4000 by 236 log-likelihood matrix.
#>
#> Estimate SE
#> elpd_loo -671.9 36.8
#> p_loo 94.4 14.5
#> looic 1343.8 73.6
#> ------
#> MCSE of elpd_loo is NA.
#> MCSE and ESS estimates assume MCMC draws (r_eff in [0.4, 2.2]).
#>
#> Pareto k diagnostic values:
#> Count Pct. Min. ESS
#> (-Inf, 0.7] (good) 230 97.5% 142
#> (0.7, 1] (bad) 5 2.1% <NA>
#> (1, Inf) (very bad) 1 0.4% <NA>
#> See help('pareto-k-diagnostic') for details.
#>
#> Output of model 'fit2':
#>
#> Computed from 4000 by 236 log-likelihood matrix.
#>
#> Estimate SE
#> elpd_loo -594.6 13.9
#> p_loo 107.2 7.1
#> looic 1189.1 27.7
#> ------
#> MCSE of elpd_loo is NA.
#> MCSE and ESS estimates assume MCMC draws (r_eff in [0.4, 1.7]).
#>
#> Pareto k diagnostic values:
#> Count Pct. Min. ESS
#> (-Inf, 0.7] (good) 178 75.4% 248
#> (0.7, 1] (bad) 55 23.3% <NA>
#> (1, Inf) (very bad) 3 1.3% <NA>
#> See help('pareto-k-diagnostic') for details.
#>
#> Model comparisons:
#> elpd_diff se_diff
#> fit2 0.0 0.0
#> fit1 -77.3 27.4Code snippets above are taken from the brms README but aren’t directly comparable.